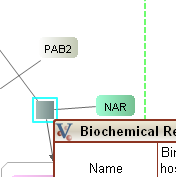
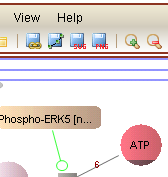
[Click on the figure to see the entire screenshot]
Pathway Visualization Services project consists of various services for effectively viewing pathway models in BioPAX format within an Internet browser.
VISIBIOweb
VISIBIOweb is a free, open-source, web-based pathway visualization and layout services software for BioPAX (level 2) pathway models. Start using now!
The user side is a thin-client JavaScript application based on Google MAPS API, whereas the server side is made up of a component based on the Eclipse Graphical Editing Framework (GEF) version 3.1.
Navigation and Inspection Facilities
With VISIBIOweb, one can obtain nicely laid out views (using specialized layout algorithms) of pathway models and can embed such views within their web pages as needed. The pathway views may be navigated through using zoom and scroll tools, and pathway objects may be inspected interactively, providing any external database references available in the data.
Following are sample screenshots from VISIBIOweb.


Below are images of pathway views produced by VISIBIOweb taken from some prominent databases. Of course, interactive inspection might be needed to better understand each view.
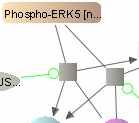
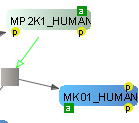
SBGN Support
VISIBIOweb complies with the standard notation of SBGN (Systems Biology Graphical Notation) for Process Diagrams (level 1). In addition, it facilitates export of such views in SBGN-ML format.
Automatic Layout Service
Automatic layout component of VISIBIOweb may also be accessed from other tools through the http protocol. The service accepts pathway graphs in a simple XML format and returns them back to the client in the same format, with layout (geometry) information added.
Software
The sources of VISIBIOweb are maintained in a sourceforge project, and distributed under Eclipse Public License. The project's Subversion repository is
A copy of the sample pathways provided by the software may be found here for download in a compressed format.
Click here to use VISIBIOweb version 1.1 (User's Guide).
Release Notes for Version 1.1
Credits
Software development team of VISIBIOweb includes Alptug Dilek, M.Esat Belviranli, Semih Ekiz, Alper Karacelik, and Ugur Dogrusoz of the Computer Engineering Department.